Amidites and Supports for 2′-O-Alkyl/Antisense Oligonucleotides:
Genes contain the information necessary to produce proteins and to understand the therapeutic and diagnostic potential of ONs, it is necessary to be aware of the mechanism of flow of genetic information from DNA to the functional proteins.
Flow of genetic information:
First DNA passes its information to messenger RNA (mRNA) by the process called ‘Transcription’. During this process, one strand of the DNA double helix is used as a template by a RNA polymerase and synthesizes premature mRNA (pre-mRNA), which contains both coding (exon) and non-coding (introns) sequences. This pre-mRNA goes through different types of maturation by the process called splicing, which involves elimination of non-coding sequences and modification at the both ends. The resulting matured mRNA, as it contains the information on protein synthesis, also called sense strand. In the final step, mRNA migrates from the nucleus to the cytoplasm, along with the ribosomes and tRNA, it synthesizes proteins, according to the genetic information forwarded by the dsDNA, by the process called translation.1, 2
Targeting of diseases - Antisense Strategies:
Proteins are vital components of all living cells and include many types of molecules necessary for carrying out the body’s functions, such as enzymes, hormones and antibodies. The overproduction or abnormal production of proteins is associated with many diseases. Traditional method to combat such diseases is to target the proteins by the small molecule drugs. In this approach small molecule drugs are designed to interact with the proteins that support or cause disease, thereby modulating its function. Binding mode of the traditional drug agents depends on the shape of proteins or charge interactions of drug molecule with the proteins. Such a binding mode has a possibility for undesired interactions with the unintended protein targets. This often leads to unexpected side effects, which serve as a major drawback of this approach.
On the other hand, oligonucleotide (ON) mediated targeting of the diseases at mRNA level or even at the genomic DNA level are promising alternatives to these traditional methods. By the former approach undesired protein synthesis is blocked by inhibiting translation (antisense strategy),3 where as by the latter approach protein synthesis is stopped at the transcription (antigene strategy).4 In these strategies diseases are fought at the earlier stages than the traditional method. Each mRNA gives rise to a relatively large number of specific protein molecules,2 which leaves with the several target molecules. Hence, inhibition of gene expression is uncomplicated than inhibition of the resulting protein product.
Conceptually, ON mediated gene silencing sounds simple and involves following steps; synthesis of complementary ON probe to the target sequence then introducing into the cell, where it binds with the target sequence (mRNA or dsDNA), thereby blocks the genetic flow of information. However, for the successful application of these strategies, several practical difficulties have to be surpassed. High affinity and specificity of an ON probe with the optimum cellular uptake, nucleolytic resistance, minimum toxicity and possibility for the mass production at a realistic price are essential requirements for the ideal antisense or antigene molecule. In addition, accessible sites of the target molecules for ON binding have to be identified. However, human genome project5 made it possible to identify the genes associated with most human diseases and determining the sequence of their genetic codes. Major efforts over the past two decades towards the search of ON probes acquiring the above mentioned properties has resulted in the satisfactory development in related synthetic methodology and chemical development. Here, I have outlined a brief description of these two strategies. Antisense (i.e. sequences complimentary to the “sense” strand, or usually messenger RNA) and otherwise interfering (e.g. “decoy”) oligonucleotides, oligoribonucleotides (ONs), and modified ONs have gained overwhelming popularity for interference in various steps leading from DNA transcription to mRNA translation. Such regulatory interference has been harnessed for therapeutic effects against many diseases and viral infections. Production of viral proteins is inhibited by oligonucleotides that are complimentary to portions of mRNA for that protein (i.e. antisense). Other regulatory mechanisms of viruses, such as HIV, have been targeted for interference with the use of complimentary (i.e. antisense), or decoy (i.e. sense) ONs. Oligonucleotides for therapy have several advantages. These include the extremely high specificity and the ease of design based on Watson-Crick base pairing. The high association constants imply a strong duplex formation and thus effectiveness at low concentrations.
Antisense strategy:
As mentioned earlier, antisense technology uses either DNA or RNA molecules that are complementary to sequences on the target mRNA and inhibits the protein production. The potential of antisense strategy as therapeutic agents was first came into picture from the discovery of the ON directed gene inhibition at the level of translation by Zamecnik and Stephenson.6 Since then, antisense technology has been developed as a potent tool for diagnostic and therapeutic purposes. The specificity of this mechanism is anticipated to develop a new class of drugs with a wide range of potential clinical applications. It is important to note that one antisense based drug, Vitravene (Fomivirsen),7 has already been approved by the US Food and Drug Administration and is used for the treatment of cytomegalovirus-induced retinitis, a disease leading to blindness that mainly affects persons with AIDS. In addition, a vast number of other modified antisense ONs (AS-ONs) are progressing in clinical phase trials and are being tested for the treatment of a wide variety of disorders.3c This serve as a major impetus for the researcher working within this field.
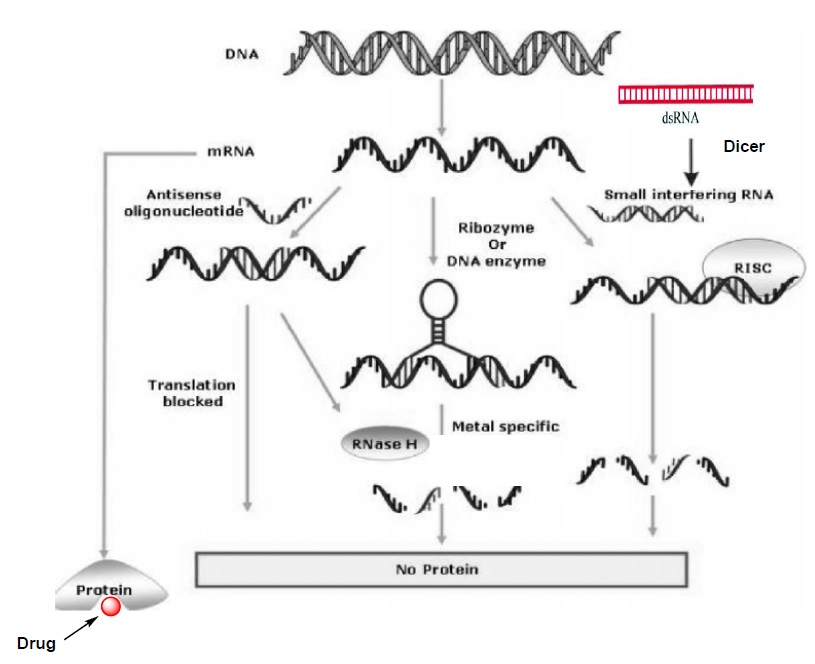
Figure 1: Illustrating the various antisense approaches along with the conventional drug therapy adopted from ref. 7b and slightly modified according to the description in my thesis.
Three different anti mRNA strategies were illustrated in Figure 1 and this figure also demonstrates the difference between antisense approaches and conventional drug therapy.3b In the first approach single stranded AS-ON probe (usually 13-25 nucleotides long) are used for the gene regulation. These AS-ONs are known to inhibit mRNA function in several ways such as, interfering with the splicing and other cellular process during the maturation of mRNA.8 However, evidence has been reported for the two major mechanisms that contribute to the antisense activity. The first and the most widely described mechanism of action is activation of enzyme RNase H, which cleaves mRNA of the AS-ON:mRNA hetero duplex and leaves the AS-ON intact.3b,c The mechanism of action is catalytic i.e., once an mRNA molecule is cleaved, the AS-ON dissociates from the duplex and becomes available to bind a second target mRNA molecule. This result in destruction of multiple mRNA targets with a single AS-ONs. RNase H is a ubiquitously expressed endonuclease and as mentioned before it recognizes a DNA:RNA heteroduplex.9 The intermediate geometry between A- and B- type conformation of hetero duplex serve as a key element for the recognition of RNase H activity, which then leads to the cleavage of the RNA strand of the heteroduplex. If the AS-ONs are incompatible with activation of RNase H, then they induce the gene silencing by the steric block approach.8 A general requirement for an AS-ON to be successful in mediating translational arrest seem to be a very high-affinity interaction of antisense probe to the mRNA target, which leads to the sterical blocking of a transcript into a protein.
In the second approach, mRNA is cleaved through catalytically active ONs referred to as ribozymes10a or DNA enzymes.10b These ON based enzymes has catalytic domain flanked by two substrate-recognition domains. The substrate domains binds to the target mRNA through Watson-Crick base pairing and catalytic core induces the site specific cleavage of phosphodiester linkage of the target mRNA, there by silences the gene (Figure 1).10 These Ribozyme and DNA enzyme with high affinity recognition arms and a partially protected catalytic domain could anticipate to enhance catalytic activity.
The third approach RNA interference (RNAi)11 is the innate cellular process that induces gene silencing by the small interfering RNA (siRNA) molecules. Since the first description of this process in the nematode C. elegans12 RNAi has become a powerful and widely used tool for gene down regulation and Andrew Fire and Craig Mello were awarded the Nobel Prize in medicine for the year 2006 in an honor of recent developments in RNAi mechanism. RNAi pathway is initiated by an endogenous ribonuclease enzyme called Dicer, which process long dsRNA molecules into short fragments of 21-23-nucleotide long siRNA molecules with the two nucleotide 3′-overhanging ends. The siRNA molecules incorporated into RNA-induced silencing complex (RISC), where one of the two siRNA strands get unwinds. The single-stranded antisense molecule that is associated in RISC, sequence selectively binds to the specific mRNA transcript and an endonuclease from this complex cleaves the target mRNA (Figure 1). From the last few years, siRNA approach has become the preferred method for gene down regulation in many academic and industrial laboratories and it holds great therapeutic application for gene silencing.13
Antisense strategy, with the above mentioned advantages, has potential to serve as a key instrument for future therapies and also a powerful tool in molecular biology for the manipulation of gene expression.
Synthetic Antisense oligo (2′-O-me Modified) are designed to a) Permeate through cell membrane, b) Bind to target RNA and c) Inactivate the target RNA.
Biological Properties of 2′-O-Methyl RNA Molecules:
- 2′-O-Methyl oligonucleotides have been used as chimeric oligomers with DNA-oligomers for site directed cleavage of RNA with RNaseH.14,15
- 2′-O-Methyl oligonucleotides are more stable against several nucleases as compared to the deoxy and ribo type of oligomers. Therefore, the 2′-O-Methyl oligonucleotides are less susceptible to several nucleases.16
- The use of 2′-O-Methyl and mixed phosphorothioates results in an inhibitory effect against HIV induced cytopathic effects and expressions of the virus specific antigens in cultures MT-4 cells. The anti-viral activity seems to be strongly concerned with the resistance to one or the other kind of deoxynucleases.17
- 2′-O-Methyl oligoribonucleotides form stable hetero-duplexes with the complimentary RNA. The hybrid formed has a high, or a higher Tm than the corresponding DNA sequence.14
- 2′-O-Methyl ribonucleotides have been used as valuable antisense probes for studying pre-mRNA splicing and the structures of splicosomes.18,19
- The 2′-O-Methylethers of common ribonucleosides have been found as minor components of RNA.20,21
- A number of biotechnology oriented companies in the U.S. are currently involved in the development of DNA-based diagnostic and therapeutic products, using 2′-O-Methyl oligonucleotide sequences,22 such as GEMR132 being developed for HCMV treatment.23
- Antisense ONS have inhibited several gene functions in mammalian cell lines such as myc, bcl-2 (relevant to lymphomas), and erb-B2 (relevant to breast cancer).
References:
1. Stryer, L. Biochemistry, 4th Ed. W. H. Freeman and Company; New York, 1995.
2. Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th Ed. Garland science, New York, 2002.
3. a) Uhlmann, E.; Oeyman, A. Chem. Rev. 1990, 90, 543-584; b) Kurreck, J. Eur. J. Biochem. 2003, 270, 1628-1644; c) Abou l-Fadl, T. Curr. Med. Chem. 2005, 12, 2193-2214.
4. Buchini, S.; Leumann, C. J. Curr. Opin. Chem. Biol. 2003, 7, 717-726.
5. a) International Human genome Sequencing Consortium, Nature 2001, 409, 860-921. b) Venter, J. C. et. al. Science, 2001, 291, 1304-1351.
6. Stephenson, M. L.; Zamecnik, P. C. Proc. Natl. Acad. Sci. USA 1978, 75, 285- 288; b) Zamecnik, P. C.; Stephenson, M. L. Proc. Natl. Acad. Sci. USA 1978, 75, 280-284.
7. a) Marwick, C. J. Am. Med. Assoc. 1998, 280, 867-871. b) Kaur, H.; Babu, B. R.; Maiti, S. Chem. Rev. 2007, 107, 4672-4697.
8. Baker, B. F.; Monia, B. P. Biochim. Biophys. Acta, 1999, 1489, 3-18.
9. Fedorof, O. Y.; Salazer, M.; Reid, B. R. J. Mol. Biol. 1993, 233, 509-523.
10. a) Doherty, E. A.; Doudna, J. A. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 457-475; b) Santoro, S. W.; Joyce, G . F. Proc. Natl. Acad. Sci. USA 1997, 94, 4262-4266.
11. Dykxhoorn, D. M., Novina, C. D.; Sharp, P.A. Nat. Rev. Mol. Cell Biol. 2003, 4, 457-467.
12. Fire, A.; Xu, S.; Montgomery, M. K.; Kostas, S. A.; Driver, S. E.; Mello, C. C. Nature 1998, 391, 806-811.
13. a) Dorsett, Y.; Tuschl, T. Nat. Rev. Drug Disc. 2004, 3, 318-329; b) Uprichard, S. L. FEBS Lett. 2005, 579, 5996-6007.
14. Inoune, H.; Hayese, Y.; Imura, A.; Iwai, S.; Miura, K.; Ohtsuka, E. Nucl. Acids Res. 1987, 15, 6131-6148.
15. Shibahara, S.; Mukai, S.; Nishihara, T.; Inoune, H.; Ohtsuka, E.; Morisawa, H. Nucl. Acids Res. 1987, 15, 4403-4415.
16. Dunlap, B. F.; Friderici, K. H.; Rottman, F. Biochemistry, 1971, 10, 2581-2587.
17. Shibahara, S.; Mukai, S.; Morisawa, H.; Nakashima, H.; Kobayashi, S.; Yamamoto, N. Nucl. Acids Res. 1989, 17, 239-252.
18. Lamond, A. I.; Sproat, B. S.; Ryder, U.; Hamm, J. Cell 1989, 58, 383-390.
19. Barbino, S.; Sproat, B. S.; Ryder, U.; Blencowe, B. J.; Lamond, A. I. Cell, 1989, 59, 531-539.
20. Hall, R. H. “The Modified Nucleosides in Nucleic Acids”, Columbia University Press, 1971, New York, NY.
21. Cotton, M.; Oberhauser, B.; Schaffner, G.; Wagner, E.; Birnsteil, M. L. Nucl. Acids Res. 1991, 19, 2629-2635.
22. Agarwal, S.; Tang, J. Y., Antisense Res. and Devel. 1992, 4, 261.
23. Michael, J. G. XII International Roundtable, Nucleosides, Nucleotides & Their Biological Applications, 1996, La Jolla, CA, Sep 15-19, Abstract 0850.


